Overview
Heart failure is growing epidemic worldwide with significant morbidity and mortality. There is need for new and more targeted therapies. Our lab is investigating novel pathways and ways to provide more targeted therapies.
Goals and Approaches
- We use molecular, biomechanical and computational approaches to study pathogenesis of acquired and genetic cardiomyopathies. We are interested in precision medicine and novel sarcomeric-based therapies.
- The experimental approaches used in our group span from recombinant proteins and isolated myosin motors to myofibrils, multicellular preparation, cells and whole organ analysis.
- Computational approaches in the lab include multi-scale model of the sarcomere and large data analytics.
Current Projects
Multi-scale computational modeling of the sarcomere to determine if a variant is pathogenic and to look for emerging twitch properties.
In collaboration with the Daniel laboratory (UW Biology), we use a spatially explicit computational model of the sarcomere to simulate how perturbations in sarcomere mechanochemistry change myocyte force generation. The spatially explicit, multi-filament computational model of the half sarcomere is a stochastic kinetic model consisting of four myosin filaments and eight actin filaments arranged in a double-hexagonal lattice. There is geometrical information, such as double helical actin filament and tropomyosin strands, with troponin spaced every 7 actins and a 2-to-1 ratio of thin:thick filaments with correctly spaced myosin head crowns projecting from thick filament backbones. The simulations are driven by experimental observations and are used as tools to predict sarcomeric variant twitch phenotypes. The model can generate large data sets of twitches that we use for data analytics.
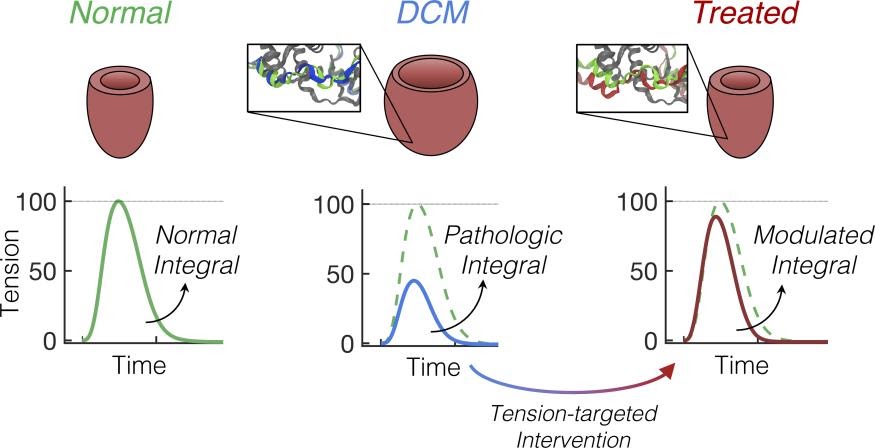
Cardiac twitch as a predictor of disease and response to therapy in cardiomyopathies.
In collaboration with Davis (UW Bioengineering/UW Lab Medicine and Pathology) and Regnier (UW Bioengineering) labs, we have shown that the tension-time index (TTI: the area of a tension-time graph) in rodent or human induced pluripotent cardiomyocytes (hiPSC-CM) is predictive of cardiomyopathy phenotype (hypertrophic or dilated). We have recently shown that correcting the abnormal TTI results in normalizing the abnormal remodeling in a mouse model of dilated cardiomyopathy. We are now working on studying effects of small molecule sarcomere modulators on correcting abnormal contractile properties in genetic cardiomyopathies. We are interested in “Precision Medicine” and predicting response to therapy in groups of sarcomeric variants.
Deoxynucleotide regulatory pathways in cardiac development, disease and as a therapeutic target
In collaboration with the Regnier lab (UW Bioengineering), we have shown that the small molecule dATP increases contractility of cardiac muscle by increasing myosin crossbridge binding and cycling. Overexpression of the enzyme Ribonucleotide Reductase (RNR) increases dATP in cardiomyocytes, resulting in stronger, faster cardiomyocyte contraction & relaxation. We are developing this as gene therapy for heart failure. Our lab has also developed a reproducible and sensitive assay to measure dATP and other deoxynucleotides at femtomole level using HPLC–MS/MS. We are also investigating the role of RNR in cardiomyocyte homeostasis using a novel mouse model with ability to specifically reduce RNR activity in cardiomyocytes.
Leadership

Principal Investigator
Farid Moussavi-Harami, MD
Farid Moussavi-Harami, MD
Assistant Professor, Medicine
John and Cookie Laughlin Endowed Professorship in Cardiology
Collaborators
- Michael Regnier, Ph.D (UW Bioengineering)
- Jennifer Davis, Ph.D (UW Bioengineering/UW Lab Medicine and Pathology)
- Thomas Daniel (UW Biology)
- Jil Tardif, MD (University of Arizona)