Cardiac Disease Modeling
Overview
Genetic heart diseases (hypertrophic, dilated, and arrhythmogenic cardiomyopathies) cause significant morbidity and mortality. While pathogenic variants in genes associated with these cardiac conditions are often medically actionable, the vast majority of the variants in these genes are variants of unknown significance (VUS), which can lead to delays in diagnosis and treatment. To help address this VUS problem, we determine the effect these variants have on cardiac diseases by studying variants at scale in cardiomyopathic genes with human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes. We are currently studying myosin heavy chain 7 (MYH7), a gene that is associated with both hypertrophic and dilated cardiomyopathy. Ongoing projects are focused on functionally characterizing all possible MYH7 single nucleotide variants and elucidating variant-specific disease mechanisms. By doing so, we hope to translate our discoveries into better diagnostics and therapeutics for patients with MYH7 cardiomyopathies.
Goals and Approaches
We aim to understand the molecular basis of genetic cardiomyopathies in order to develop better diagnostic tools and novel therapies. Our approaches include:
- Modeling genetic cardiac diseases with human iPSC-derived cardiomyocytes to elucidate disease mechanisms.
- Gene-editing variant libraries into hiPSC-derived cardiomyocytes at scale, such that each cardiomyocyte expresses a different variant.
- Functionally annotating all possible MYH7 variants in hiPSC-derived cardiomyocytes with high-throughput assays.
Current Projects
High-throughput characterization of variants of unknown significance
Variants of unknown significance (VUS) present a challenge for clinical geneticists and patients. Discovery of a VUS in a patient with myocardial disease can have an emotional impact on patients and family members and often leads to further testing of significant cost and uncertain value. At worse, it may lead to a missed opportunity for early diagnosis and therapeutic intervention. Pathogenic variants in the myosin heavy chain 7 gene (MYH7) are among the most common causes of genetic cardiomyopathies; however, thousands of MYH7 variants are classified as VUS in the publicly available database, ClinVar. We combine gene-editing techniques with high-throughput functional assays, with an ultimate goal of classifying all MYH7 VUS as pathogenic or not. Patients with pathogenic variants will merit further surveillance and treatment whereas patients with benign variants can be reassured.
Mechanistic studies in human iPSC-derived cardiomyocytes
Because sarcomeric mutations can lead to the accumulation of toxic myocardial proteins known as “poison peptides,” we are interested in defining cellular pathways that regulate clearance of sarcomeric proteins. Our long-term goal is to apply this knowledge to develop drug therapy that would improve cardiomyocyte function and prevent disease caused by pathogenic sarcomeric variants. To achieve this, we perturb the ubiquitin-proteasome pathways that regulate sarcomeric proteins and measure the effect on cardiomyocyte contractility in single cells (traction force microscopy) and 3D cardiac organoids expressing different pathogenic variants. By determining variant-specific mechanisms, we hope to leverage this knowledge to develop novel therapeutics.
Research Leadership

Principal Investigator
Kai-Chun (Daniel) Yang, MD
Assistant Professor, Medicine
Email: kcyang@uw.edu
Faculty Profile: Daniel Yang, MD
Yang Lab Members
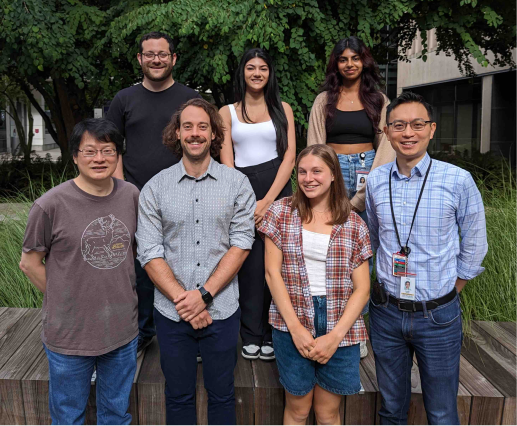
Front row left to right: Wei-Ming Chien (research scientist), Clayton Friedman (postdoc), Ashley Mckinstry (undergrad), Daniel Yang
Positions available
Postdoctoral positions are available for highly motivated individuals. If interested, please apply with the link (https://ap.washington.edu/ahr/position-details/?job_id=145199), or clicking the button below.